Assignment 1: Which variant of which gene predicts a positive prognosis in colorectal cancer
During this assignment, we will have a closer look at an example SPARQL query of Wikidata, called “Which variant of which gene predicts a positive prognosis in colorectal cancer”. We will first go through the basics of a SPARQL query. Second, we will find out how to execute the query and retain or share results. Last, we will expand the query and make other (small) changes, to understand the structure of a SPARQL query better, and see what other data is available in Wikidata.
What goes Where
A SPARQL query consist out of several elements, which can be considered as building blocks. We will use the following example, which is part of the example page of SPARQL queries for Wikidata. We will exploring the example question called “Which variant of which gene predicts a positive prognosis in colorectal cancer” in more detail below.
First element: SELECT
The first element we encouter in this example, is the so-called result clause, which identifies what information to return from the query. This element starts with the word SELECT, and is then followed by two words with a questionmark in front of them:
SELECT ?geneLabel ?variantLabel
SELECT is used to indicate with variables from the (to follow) SPARQL query you want to visualise as a result (in other words: which variables we find relevant as output to answer our biological question). In this example, the name(s) of the gene(s) predicting a positive prognosis in colorectal cancer (?geneLabel), and the name of the variant(s) belonging to this gene/these genes (?variantLabel).
Second element: WHERE
The second element we encouter, is the query pattern, which starts with the word WHERE, with the query itself enclosed in curly brackets: {} .
WHERE
{
query
}
Variable Names
Within these brackets, the data from an RDF is connected to variable names. We already encoutered two variable names, ?geneLabel and ?variantLabel (indicated by the questionmark). These variables can have any name you see fit.
Question1: Which other variable names are present in the following query? (Answers can be found here).
{
VALUES ?disease {wd:Q188874}
?variant wdt:P3358 ?disease ; # P3358 Positive prognostic predictor
wdt:P3433 ?gene . # P3433 biological variant of
}
Query Details
In between the curly brackets in the query above, we can see three lines to describe the query we want to execute:
- VALUES ?disease {wd:Q188874}
- ?variant wdt:P3358 ?disease ; # P3358 Positive prognostic predictor
- wdt:P3433 ?gene . # P3433 biological variant of
Line 1
The first line starts with VALUES; this allows you to queries multiple items, specified in the data block between the brackets at the end of this line. Multiple items should be separated with spaces. The variable after VALUES (in this case ?disease) will be completed with the items in the data block. In this case, we are only interested in “colorectal cancer”, with ‘Q188874’ being the identifier of the entry in Wikidata (these always start with a Q) for Colon Cancer. To explain to which database this identifier belongs, we add the ‘wd:’ before the number of the identifier.
Line 2
The second line consists of two main parts, the left part is related to the query (which ends with a semicolon ‘;’ ), the right is a comment, to explain what the line does (starting with a hash ‘#’). The left part is formed in the triplet structure (which we previously encountered in the presentation):
{
?variant wdt:P3358 ?disease ;
}
In this case, we are looking for variant(s) (subject), which are related to disease(s) (object). The relationship (aka predicate) is defined as the center part of the triplet, with ‘P3358’ being the identifier of the property in Wikidata (these always start with a P) for Positive prognostic predictor. To explain to which database this relationship belongs, we add the ‘wdt:’ before the identifier of the property. Notice the small difference with line one; ‘wd:’ is used for entries (these can serve as subjects or objects), ‘wdt:’ for properties (or relationships).
This line ends with a semicolon ‘;’ to end our first triplet.
Line 3
The third line again consists out of two parts; we will only discuss the left had side, until the point ‘.’ .
{
wdt:P3433 ?gene .
}
Even though this line does not directly look like a triplet (since there are only two element), it is read as a triplet. A point ‘.’ at the end of a triplet really notes the end, while a semicolon ‘;’ notes that another triplet will follow, where the subject may be ommited from writing. Therefore, we need to look back at line 2, to find out which subject is belonging to the triplet in line 3: ?variant. This line of the SPARQL query is therefore read as:
{
?variant wdt:P3433 ?gene .
}
In this case, we are defining how genes are related to the variants in Wikidata. We are connecting the variant(s) (subject) with genes (object) in Wikidata, with the following relationship(predicate): “P3433”, which is also known as “biological variant of”.
Third element: SERVICE
Note that the ?geneLabel and ?variantLabel are not written down in the query, however the variables ?gene and ?variant are. This is possible with the last part of the query, the SERVICE element. By typing the word “Label” (including the capital!) behind the name of a variable, we can obtain the name of that variable, in stead of the identifier used by the RDF. A name makes much more sense to us humans, and allows us to interpret the results.
{
SERVICE wikibase:label { bd:serviceParam wikibase:language "[AUTO_LANGUAGE],en" }
}
Additional Remark: the SERVICE clause is not default SPARQL behaviour; it is part of the Wikidata SPARQL structure (like some of the visualisation options you see later on). Therefore, this statement will (most likely) not work when building a query in any other database. The actual SPARQL query to retrieve labels without using SERVICE is explained here
Full query
When we combine the three elements above, we get the full query:
SELECT ?geneLabel ?variantLabel
WHERE
{
VALUES ?disease {wd:Q188874}
?variant wdt:P3358 ?disease ; # P3358 Positive prognostic predictor
wdt:P3433 ?gene . # P3433 biological variant of
SERVICE wikibase:label { bd:serviceParam wikibase:language "[AUTO_LANGUAGE],en" }
}
We wanted an answer for the following question: “Which variant of which gene predicts a positive prognosis in colorectal cancer”.
Question 2A: Which item in the SPARQL query corresponds to the disease(s) being queried?
Question 2B: Which item in the SPARQL query adds a name to the results?
(Answers can be found here).
We will now look how to run this query on the data from Wikidata, and how we can save the results from that query.
Run and Save
Click on this link to go to the example page of Wikidata. Below the Query titled “Which variant of which gene predicts a positive prognosis in colorectal cancer”, click on the “Try it” button, which will open the following page:
Welcome to the SPARQL Endpoint of Wikidata!
Excecute the query by clicking on the blue play button. This will reveal the results of the query in a panel below the query editor:
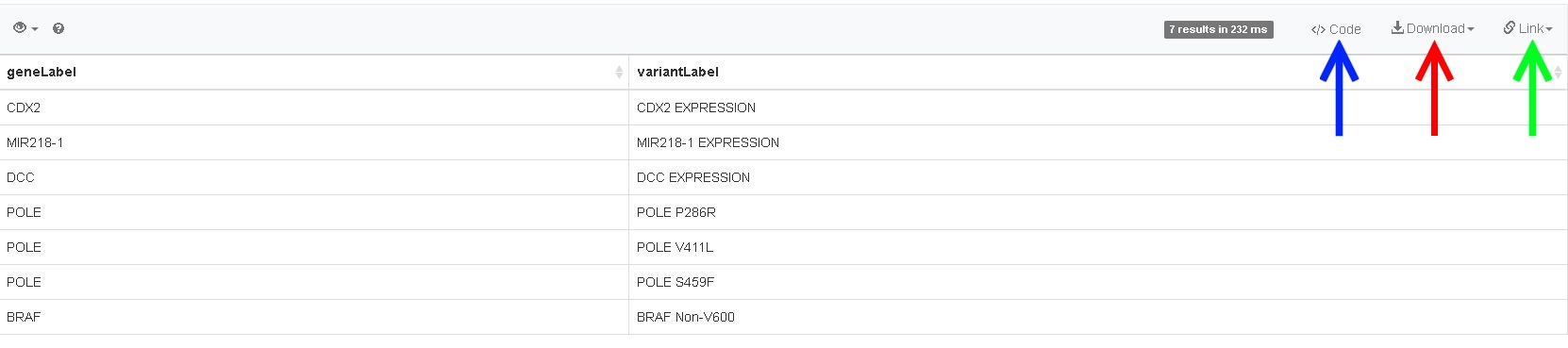
There are several options to work with the results of your query. To save your data, click on the Download button (red arrow in image below), and select the format you want to work with (CSV, TSV, JSON, HTML, SVG-image). To get a weblink to your results, click on the Link button (green arrow in image below). Last, there are also several code examples available (blue arrow in the image below), which could help construct a script to automate (several) queries, or combine the results of multiple queries in a workflow. Examples are available for: R, Python, Ruby, Perl, Java, JavaScript and many others!
We will now make some changes to this query, to understand the structure of SPARQL even better.
Change is Coming
More diseases:
We have now limited ourselves to only one disease, colorectal cancer. If we would like to add another disease, such as “breast cancer” (Q128581), we would need change the VALUES line in our query (original below):
{
VALUES ?disease {wd:Q188874}
}
By adding the identifier (Q128581) for the Wikidata entry called “breast cancer” to the VALUES element, we can expand our query to include two diseases. Change the line depicted above in the SPARQL endpoint to the following and click the blue run button again:
{
VALUES ?disease {wd:Q188874 wd:Q128581}
}
You should now see more results, compared to our previous endeavour.
Question 3: How will the line above look, when we also want to add stomach carcinoma (Q3658392) to our list?
(Answers can be found here).
Which diseases?
Since we are obtaining more results by adding more diseases to our query, it would be great if we know to which disease which variant is related. In order to obtain the disease in the results, we should change the result clause section of our SPARQL query:
SELECT ?geneLabel ?variantLabel ?disease
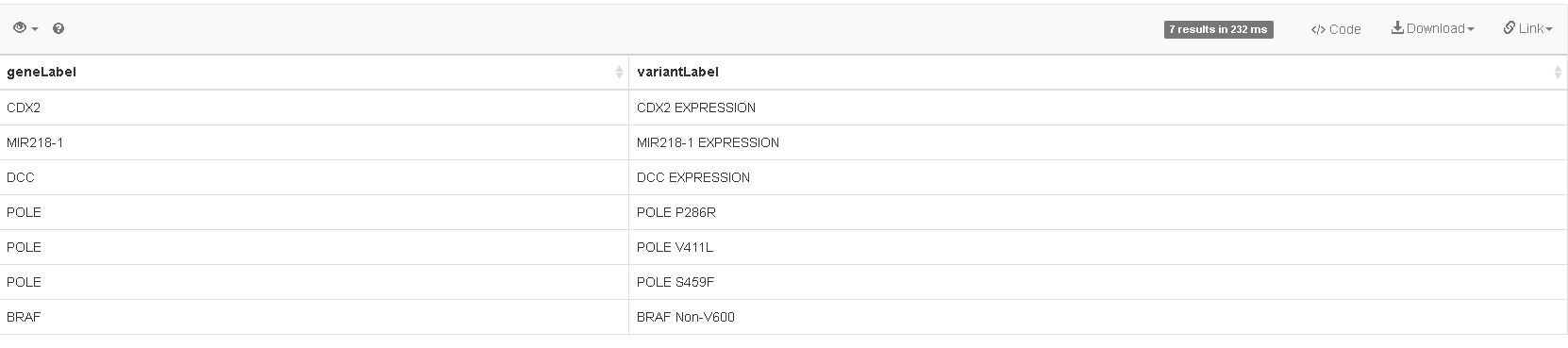
Click the play button again; there should now be three columns in your results panel… However, the disease column is only giving us the identifier from Wikidata, not the name of the disease.
Question 4: How should the line above look, when we want to see the name of the disease in our results panel?
(Answers can be found here).
Easier querying: Adding diseases with entry search function
Finding the identifiers for each entry you are interested in, can be done very easily with the entry search function. If we would like to add the disease “ovarian cancer” to our list of diseases of interest, we could do the follwing:
- In the SPARQL endpoint, find the VALUES line.
- Click just before the last curly bracket ‘}’ .
- Type a space ‘ ‘, and then ‘wd:’ .
-
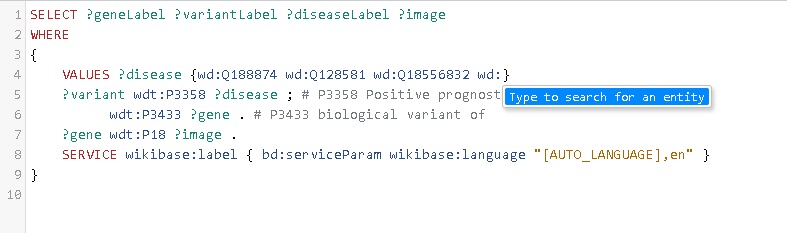
Now hit Ctrl and the spacebar on your keyboard simultaneously (Windows, for Apple: CMD in stead of Ctrl). This should open up the entry search field of Wikidata (see image below).
-
Type the words ‘ovarian cancer’ in the search field, which should trigger a search in all entries in Wikidata (see image below).
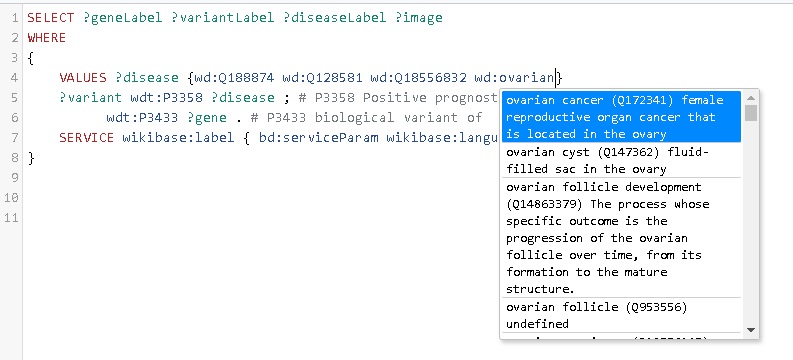
- Click on the entry with identifier Q172341; this adds the identifier to your list of VALUES.
- Run your query again.
Adding protein images
The SPARQL endpoint of Wikidata has several interesting data visualisation options; we will use one to add protein domain images for the genes we just queried.
- Just above the SERVICE element, add the following line:
?gene wdt:P18 ?image .
- Click on the play button… What just happened? We had 13 variants, and now the results went down to 6?!
Since not all genes have an image in Wikidata, we are only retrieving the ones that have an image. This can be avoided by using an OPTIONAL statement, such as:
OPTIONAL{?gene wdt:P18 ?image }.
However, we are not seeing the images in our results panel. Every time we want to see a variable that we are querying, we need to add it to the SELECT statement.
- Change the SELECT statement to the following:
SELECT ?geneLabel ?variantLabel ?diseaseLabel ?image - Click on the play button… We do not see the images directly, we do get a link to the images in a Table. If we want to actually see the images, we need to change the visualisation options of the SPARQL endpoint.
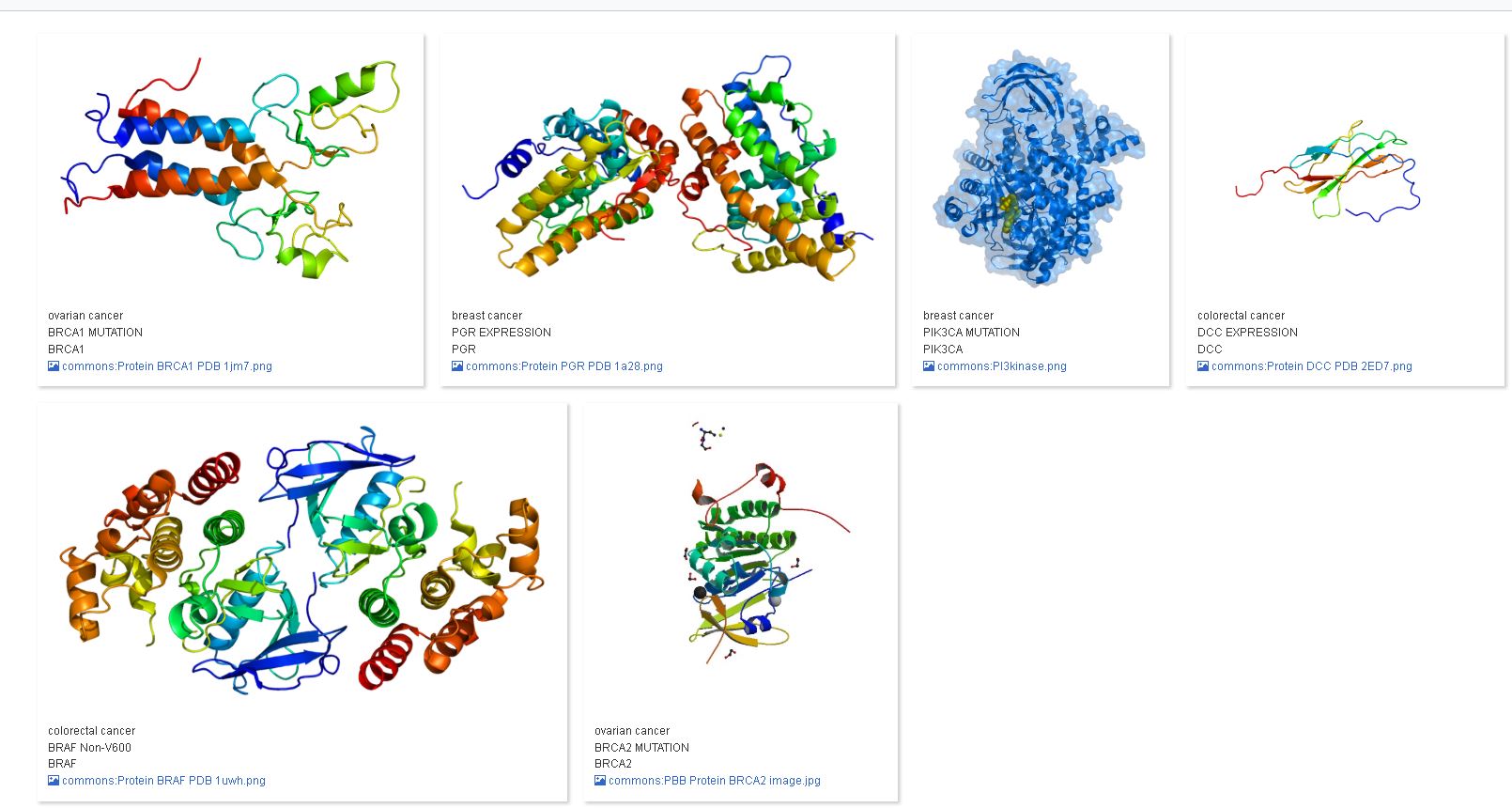
- Directly under the Run button, there is an option called ‘Table”. Click on this option, and select the option “Image Grid”:
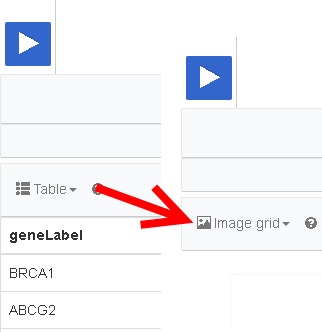
Now, the genes in Wikidata which have an image connected to them, are displayed.
If you would like to have images for all the genes you queried, you can add these to Wikidata yourself. Since the data in Wikidata is built by community efforts, everyone can get involved. If you would like to know more about becoming a database editor and/or curator for Wikidata, ask one of the instructors for more information.
Next assignments:
You can now either progress to Assignment 2, where we will discuss another query in more detail, or stay with the current query to adapt it to your own needs. Several example questions to work on are given below.
Additional questions Assignment 1:
?Negatives
We have only been looking at variant in genes which predict a positive diagnosis. There are also variants known related to a negative diagnosis. Change the query to obtain these variants.
(Answers can be found here).
?References
Finding literature that is related to genetic variants can be quite tricky in regular search engines. Obtain the names of the articles which describe these variants as “Main topic”(relationship). Change the example query to obtain the required results.
(Answers can be found here).
?MORE biology
You can probably think of some other questions you would like to ask to Wikidata. Try to come up with a expansion of the query we are working on now, or find another example on Wikidata you want to understand and expand (ask your instructors for help if needed).