PRA3006-SPARQL
WikiPathways
| License | CCZero |
WikiPathways is a database with machine-readable models of biological processes for human and multiple other species [1,2]. It comes with a SPARQL endpoint with a human-oriented interface at sparql.wikipathways.org [3].
WikiPathways RDF has two parts. The first is the GPMLRDF which is an RDF representation of the Graphical Pathway Markup Language (GPML) in which the biological pathways are stored in the database. The second is the WPRDF which is the represented biological knowledge [3,4]. This chapter focuses on the WPRDF only.
Figure of simplified RDF schema:
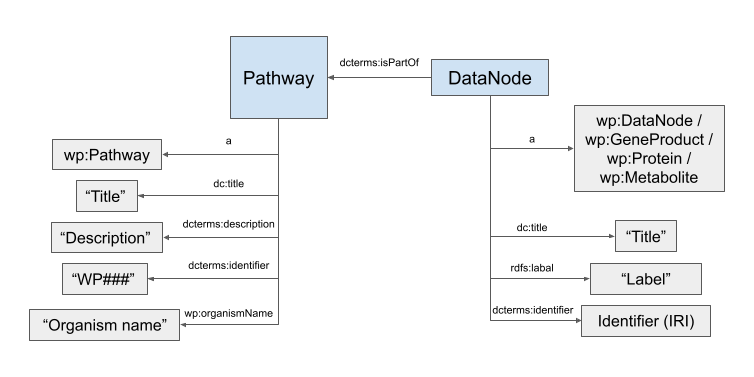
Entities
The RDF contains all pathways, their datanodes (genes, proteins, metabolites, etc.), author information, molecular descriptors, and more. The main classes are:
- Pathway: a biological pathway
- GeneProduct: can be a gene, strand of RNA, and a protein.
- Rna: RNA, e.g. miRNA.
- Protein: a protein. Post-translational modifications can be indicated with states
- Metabolite: metabolites, ions, and other small molecules. It includes peptides.
- Interaction: can be a lot of things: translocation, inhibition, metabolic conversions (see [4]).
In all cases, the specific meaning is not clearly defined. Each of the above types is roughly defined by the database identifies linked to the entity. For example, a UniProt identifier linked to a GeneProduct suggests the entity is actually a protein.
Data model
Because the WikiPathways RDF contains many properties of all subjects (such as pathways), we can also directly request all
contents through the SPARQL query. For example, to extract the pathway title, we add ?pathway dc:title ?pathwaytitle
to the SPARQL query and add ?pathwaytitle in the SELECT list. The returned table upon running the query will get
wider, so you might need to scroll to the right to see it all.
Example queries
The simplest SPARQL queries to explore RDF is to retrieve full lists of subjects of a particular type, which is
frequently defined with the predicate rdfs:type or a which can be used interchangably. See the below example
of listing all pathways.
SPARQL sparql/pathways.rq (run, edit)
SELECT ?pathway
WHERE {
?pathway a wp:Pathway .
}
The list is long and this is the first five:
| pathway | |
| https://identifiers.org/wikipathways/WP5382_r129730 | |
| https://identifiers.org/wikipathways/WP5472 | |
| https://identifiers.org/wikipathways/WP5473 | |
| This table is truncated. See the full table at sparql/pathways.rq | |
Asking information for a specific pathway
With this exercise, the RDF will be explored a little more extensively. By combining statements in the RDF query,
we can link multiple subjects and filter for content that we want to get back from the service. Important: when
filtering for a literal (gene label, organism, etc.) the literal should have the following format:
"text"^^xsd:string. For example, the next query returns the title for pathway with ID WP4846:
SPARQL sparql/pathwayWP4846.rq (run, edit)
SELECT ?pathwaytitle WHERE{
?pathway a wp:Pathway .
?pathway dc:title ?pathwaytitle .
?pathway dcterms:identifier "WP4868"^^xsd:string .
}
Which returns the following title:
A lipid pathway
For example, we can ask a list of pathways describing the biology of oxygenated hydrocarbons (LMFA12):
SPARQL sparql/lipidPathways.rq (run, edit)
PREFIX chebi: <http://purl.obolibrary.org/obo/chebi/>
SELECT ?lipid ?name ?formula ?lmid (GROUP_CONCAT(?wpid_;separator=", ") AS ?pathway)
WHERE {
SERVICE <https://lipidmaps.org/sparql> {
VALUES ?category { <https://www.lipidmaps.org/rdf/category/112> <https://www.lipidmaps.org/rdf/category/11200> }
?lipidmaps rdfs:label ?name ;
rdfs:subClassOf* ?category ;
chebi:formula ?formula .
}
BIND (IRI(CONCAT("https://identifiers.org/lipidmaps/",
SUBSTR(STR(?lipidmaps), 31))) AS ?lmid)
?lipid wp:bdbLipidMaps ?lmid ;
dcterms:isPartOf ?pathway .
?pathway a wp:Pathway ; dcterms:identifier ?wpid_ .
}
This gives:
| lipid | name | formula | lmid | pathway |
| https://identifiers.org/kegg.compound/C00207 | Propan-2-one | C3H6O | https://identifiers.org/lipidmaps/LMFA12000057 | WP3602 |
| https://identifiers.org/chebi/CHEBI:15347 | Propan-2-one | C3H6O | https://identifiers.org/lipidmaps/LMFA12000057 | WP5175, WP4742 |
| https://identifiers.org/chebi/CHEBI:28398 | Butan-2-one | C4H8O | https://identifiers.org/lipidmaps/LMFA12000043 | WP4838 |
A federated SPARQL query
This final example adds an extra level of difficulty by linking the AOP-Wiki RDF with another database through SPARQL (this is called a Federated SPARQL query). In this exercise we will explore the connection between WikiPathways and AOP-Wiki (see this chapter).
The SPARQL query will need to contain a SERVICE function and the final query will have the following structure:
PREFIX aopo: <http://vocabularies.wikipathways.org/wp#>
SELECT [variables] WHERE {
[query WikiPathways]
SERVICE <https://aopwiki.rdf.bigcat-bioinformatics.org/sparql> {
[query AOP-Wiki]
}
}
References
- Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008 Jul 22;6(7):e184. doi:10.1371/JOURNAL.PBIO.0060184 (Scholia)
- Martens M, Ammar A, Riutta A, Waagmeester A, Slenter D, Hanspers K, et al. WikiPathways: connecting communities. NAR [Internet]. 2021 Jan 8;49(D1):D613–21. Available from: https://academic.oup.com/nar/article/49/D1/D613/5992285 doi:10.1093/NAR/GKAA1024 (Scholia)
- Waagmeester A, Summer-Kutmon M, Riutta A, Miller R, Willighagen E, Evelo CT, et al. Using the Semantic Web for Rapid Integration of WikiPathways with Other Biological Online Data Resources. PLoS Comput Biol. 2016 Jun;12(6):e1004989. doi:10.1371/JOURNAL.PCBI.1004989 (Scholia)
- Miller RA, Kutmon M, Bohler A, Waagmeester A, Evelo CT, Willighagen EL. Understanding signaling and metabolic paths using semantified and harmonized information about biological interactions. PLOS ONE. 2022 Apr 18;17(4):e0263057. doi:10.1371/JOURNAL.PONE.0263057 (Scholia)